Reshaping Data
Last updated on 2023-04-27 | Edit this page
Estimated time: 25 minutes
Overview
Questions
- How can change the shape of my data?
- What is the difference between long and wide data?
Objectives
- Extract substrings of interest.
- Format dynamic strings using f-strings.
- Explore Python’s built-in string functions
Key Points
- Strings can be indexed and sliced.
- Strings cannot be directly altered.
- You can build complex strings based on other variables using f-strings and format.
- Python has a variety of useful built-in string functions.
Wide data and long data
We’ve so far seen our gene expression dataset in two distinct formats which we used for different purposes.
Our original dataset contains a single count value per row, and has all metadata included:
PYTHON
import pandas as pd
url = "https://raw.githubusercontent.com/ccb-hms/workbench-python-workshop/main/episodes/data/rnaseq_reduced.csv"
rnaseq_df = pd.read_csv(url)
print(rnaseq_df)OUTPUT
gene sample expression time
0 Asl GSM2545336 1170 8
1 Apod GSM2545336 36194 8
2 Cyp2d22 GSM2545336 4060 8
3 Klk6 GSM2545336 287 8
4 Fcrls GSM2545336 85 8
... ... ... ... ...
7365 Mgst3 GSM2545340 1563 4
7366 Lrrc52 GSM2545340 2 4
7367 Rxrg GSM2545340 26 4
7368 Lmx1a GSM2545340 81 4
7369 Pbx1 GSM2545340 3805 4
[7370 rows x 4 columns]
Data in this format is referred to as long data.
We also looked at a version of the same data which only contained expression data, where each sample was a column:
PYTHON
url = "https://raw.githubusercontent.com/ccb-hms/workbench-python-workshop/main/episodes/data/expression_matrix.csv"
expression_matrix = pd.read_csv(url, index_col=0)
print(expression_matrix.iloc[:10, :5])OUTPUT
GSM2545336 GSM2545337 GSM2545338 GSM2545339 GSM2545340
gene
AI504432 1230 1085 969 1284 966
AW046200 83 144 129 121 141
AW551984 670 265 202 682 246
Aamp 5621 4049 3797 4375 4095
Abca12 5 8 1 5 3
Abcc8 2210 1966 2181 2362 2475
Abhd14a 490 495 474 468 489
Abi2 5627 4383 4107 4062 4289
Abi3bp 807 1144 1028 935 879
Abl2 2392 2133 1891 1645 1926Data in this format is referred to as wide data.
Each format has its pros and cons, and we often need to switch between the two. Wide data is more human readable, and allows for easy comparision across different samples, such as finding the genes with the highest average expression. However, wide data requires any sample metadata to exist as a separate dataframe. Thus it is much more difficult to examine sample metadata, such as calculating the mean expression at a particular timepoint.
In contrast, long data is considered less human readable. We have to first aggregate our data by gene if we want to do something like calculate average expression. However, we can easily group the data by whatever sample metadata we wish. We will also see in the next session that long data is the preferred format for plotting.
Wide and long data also handle missing data differently. In long
data, and values which don’t exist for a particular sample simply aren’t
rows in the dataset. Thus, our data does not have null
values, but it is harder to tell where data is missing. In wide data,
there is a matrix position for every combination of sample and gene.
Thus, any missing data is shown as a null value. This can
be more difficult to deal with, but makes missing data clear.
Convert from wide to long with melt.
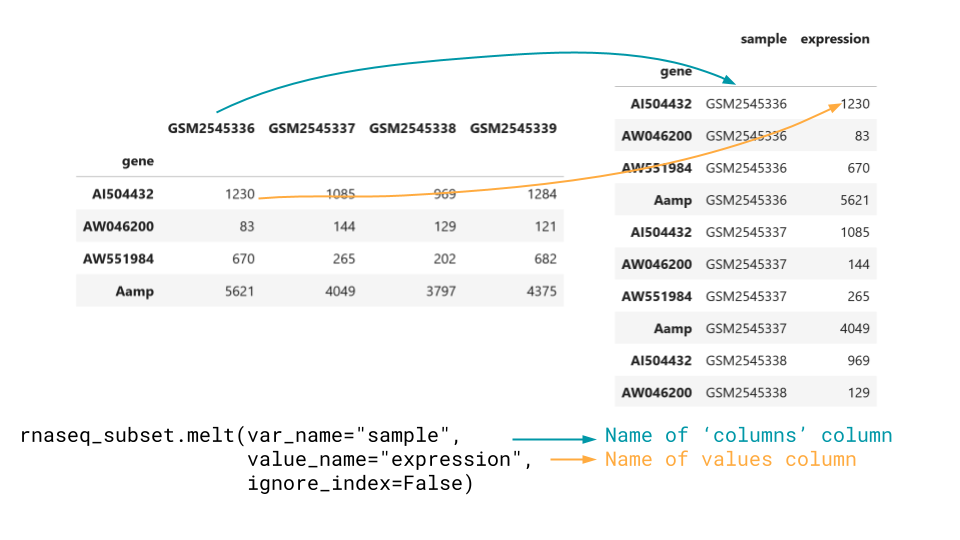
We can convert from wide data to long data using melt.
melt takes in the arguments var_name and
value_name, which are strings that name the two created
columns. We also typically want to set the ignore_index
argument to False, otherwise pandas will drop the dataframe
index.
We could also allow pandas to drop the index, if there is some other
column we want the new rows to be named by, and pass that in instead as
the id_vars argument.
PYTHON
long_data = expression_matrix.melt(var_name="sample", value_name="expression", ignore_index=False)
print(long_data)OUTPUT
sample expression
gene
AI504432 GSM2545336 1230
AW046200 GSM2545336 83
AW551984 GSM2545336 670
Aamp GSM2545336 5621
Abca12 GSM2545336 5
... ... ...
Zkscan3 GSM2545380 1900
Zranb1 GSM2545380 9942
Zranb3 GSM2545380 202
Zscan22 GSM2545380 527
Zw10 GSM2545380 1664
[32428 rows x 2 columns]Convert from long to wide with pivot.
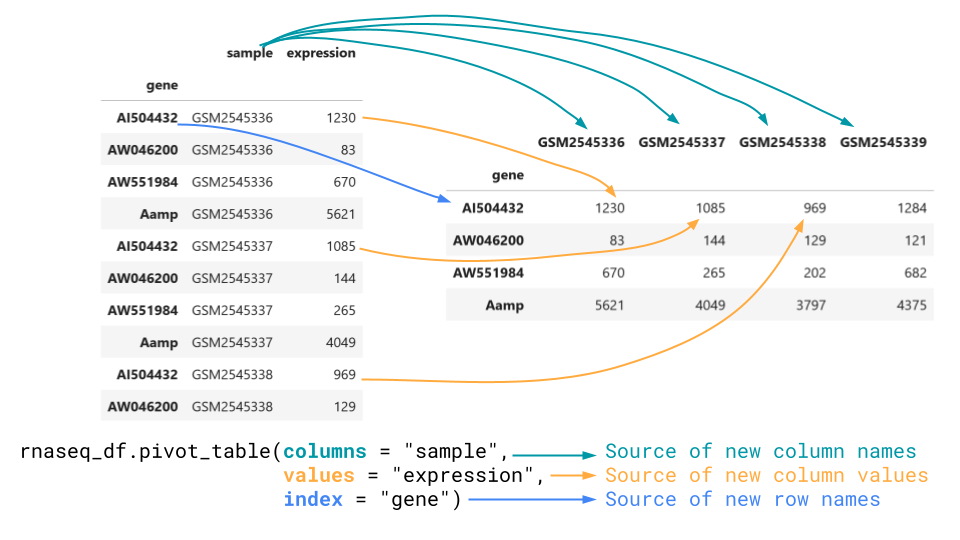
To go the other way, we want to use the pivot_table method.
This method takes in a columns the column to get new
column names from, values, what to populate the matrix
with, and index, what the row names of the wide data should
be.
PYTHON
wide_data = rnaseq_df.pivot_table(columns = "sample", values = "expression", index = "gene")
print(wide_data)OUTPUT
sample GSM2545336 GSM2545337 GSM2545338 GSM2545339 GSM2545340
gene
AI504432 1230 1085 969 1284 966
AW046200 83 144 129 121 141
AW551984 670 265 202 682 246
Aamp 5621 4049 3797 4375 4095
Abca12 5 8 1 5 3
... ... ... ... ... ...
Zkscan3 1732 1840 1800 1751 2056
Zranb1 8837 5306 5106 5306 5896
Zranb3 207 179 199 208 184
Zscan22 483 535 533 462 439
Zw10 1479 1394 1279 1376 1568
[1474 rows x 5 columns]Note that any columns in the dataframe which are not used as values are dropped.
Challenge
Create a dataframe of the rnaseq dataset where each row is a gene and each column is a timepoint instead of a sample. The values in each column should be the mean count across all samples at that timepoint.
Take a look at the aggfunc argument in pivot_table.
What is the default?
