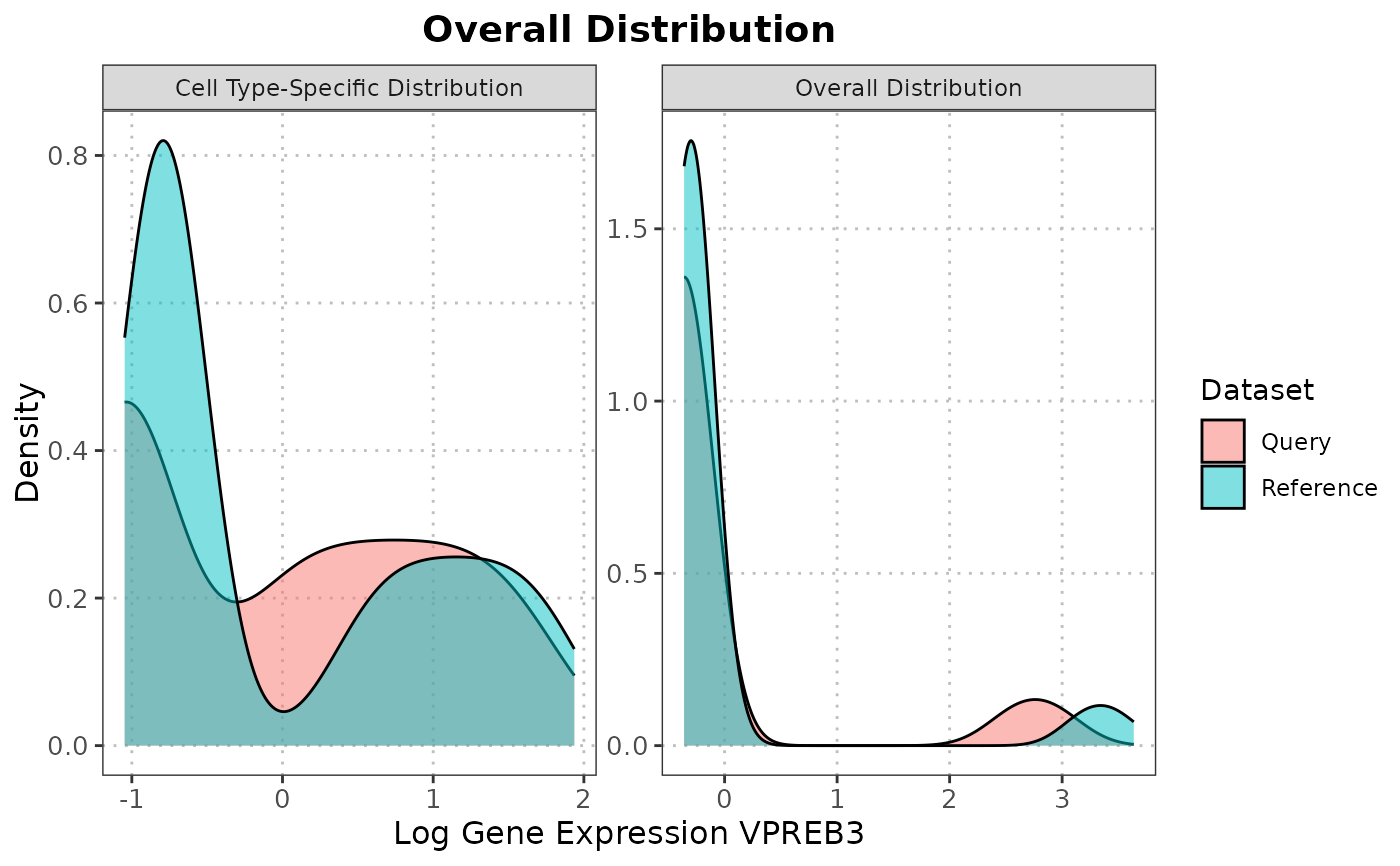
Plot gene expression distribution from overall and cell type-specific perspective
Source:R/plotMarkerExpression.R
plotMarkerExpression.RdThis function generates density plots to visualize the distribution of gene expression values for a specific gene across the overall dataset and within a specified cell type.
plotMarkerExpression(
query_data,
reference_data,
ref_cell_type_col,
query_cell_type_col,
cell_type,
gene_name,
assay_name = "logcounts",
normalization = c("z_score", "min_max", "rank", "none"),
max_cells_query = NULL,
max_cells_ref = NULL
)Arguments
- query_data
A
SingleCellExperimentobject containing numeric expression matrix for the query cells.- reference_data
A
SingleCellExperimentobject containing numeric expression matrix for the reference cells.- ref_cell_type_col
The column name in the
colDataofreference_datathat identifies the cell types.- query_cell_type_col
The column name in the
colDataofquery_datathat identifies the cell types.- cell_type
A cell type to plot (e.g., c("T-cell", "B-cell")).
- gene_name
The gene name for which the distribution is to be visualized.
- assay_name
Name of the assay on which to perform computations. Default is "logcounts".
- normalization
Method for normalizing expression values. Options: "z_score" (default), "min_max", "rank", "none".
- max_cells_query
Maximum number of query cells to retain after cell type filtering. If NULL, no downsampling of query cells is performed. Default is NULL.
- max_cells_ref
Maximum number of reference cells to retain after cell type filtering. If NULL, no downsampling of reference cells is performed. Default is NULL.
Value
A ggplot object containing density plots comparing reference and query distributions.
Details
This function generates density plots to compare the distribution of a specific marker gene between reference and query datasets. The aim is to inspect the alignment of gene expression levels as a surrogate for dataset similarity. Similar distributions suggest a good alignment, while differences may indicate discrepancies or incompatibilities between the datasets.
Multiple normalization options are available: - "z_score": Standard z-score normalization within each dataset - "min_max": Min-max scaling to [0,1] range within each dataset - "rank": Maps values to quantile ranks (0-100 scale) - "none": No transformation (preserves original scale differences)
Examples
# Load data
data("reference_data")
data("query_data")
# Note: Users can use SingleR or any other method to obtain the cell type annotations.
plotMarkerExpression(reference_data = reference_data,
query_data = query_data,
ref_cell_type_col = "expert_annotation",
query_cell_type_col = c("expert_annotation", "SingleR_annotation")[1],
gene_name = "CD8A",
cell_type = "CD4",
normalization = "z_score")
#> Picking joint bandwidth of 0.303
#> Picking joint bandwidth of 0.234