Dictionaries
Last updated on 2023-04-20 | Edit this page
Overview
Questions
- How is a dictionary defined in Python?
- What are the ways to interact with a dictionary?
- Can a dictionary be nested?
Objectives
- Understanding the structure of a dictionary.
- Accessing data from a dictionary.
- Practising nested dictionaries to deal with complex data.
Key Points
- Dictionaries associate a set of values with a number of keys.
- keys are used to access the values of a dictionary.
- Dictionaries are mutable.
- Nested dictionaries are constructed to organise data in a hierarchical fashion.
- Some of the useful methods to work with dictionaries are: .items(), .get()
Dictionary
One of the most useful built-in tools in Python, dictionaries associate a set of values with a number of keys.
Think of an old fashion, paperback dictionary where we have a range of words with their definitions. The words are the keys, and the definitions are the values that are associated with the keys. A Python dictionary works in the same way.
Consider the following scenario:
Suppose we have a number of protein kinases, and we would like to associate them with their descriptions for future reference.
This is an example of association in arrays. We may visualise this problem as displayed below:
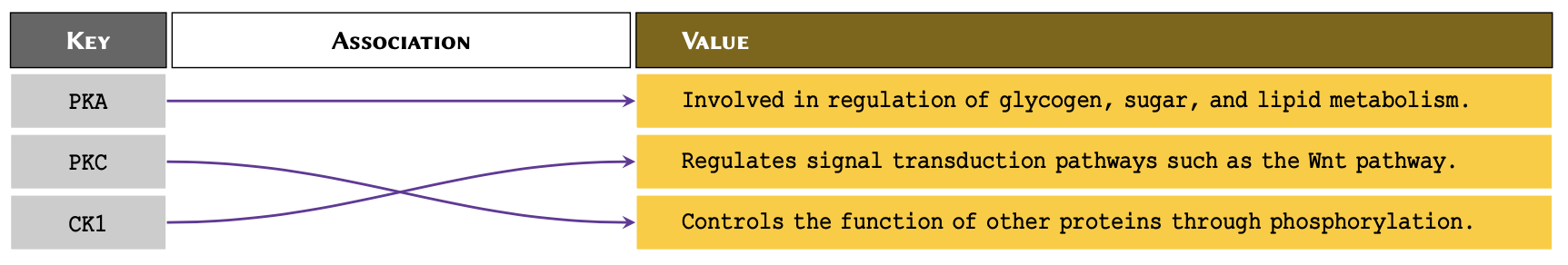
One way to associate the proteins with their definitions would be to use nested arrays. However, it would make it difficult to retrieve the values at a later time. This is because to retrieve the values, we would need to know the index at which a given protein is stored.
Instead of using normal arrays, in such circumstances, we use
associative arrays. The most popular method to create construct
an associative array in Python is to create dictionaries or
dict.
Remember
To implement a dict in Python, we place our entries in
curly bracket, separated using a comma. We separate keys and
values using a colon — e.g. {‘key’: ‘value’}. The combination of
dictionary key and its associating value is known as a
dictionary item.
Note
When constructing a long dict with several
items that span over several lines, it is not necessary to
write one item per line or use indentations for each
item or line. All we must is to write the as {‘key’: ‘value’} in curly brackets and
separate each pair with a comma. However, it is good practice to write
one item per line and use indentations as it makes it
considerably easier to read the code and understand the hierarchy.
We can therefore implement the diagram displayed above in Python as follows:
PYTHON
protein_kinases = {
'PKA': 'Involved in regulation of glycogen, sugar, and lipid metabolism.',
'PKC': 'Regulates signal transduction pathways such as the Wnt pathway.',
'CK1': 'Controls the function of other proteins through phosphorylation.'
}
print(protein_kinases)OUTPUT
{'PKA': 'Involved in regulation of glycogen, sugar, and lipid metabolism.', 'PKC': 'Regulates signal transduction pathways such as the Wnt pathway.', 'CK1': 'Controls the function of other proteins through phosphorylation.'}OUTPUT
<class 'dict'>Constructing dictionaries
Use Universal Protein Resource (UniProt) to find the following proteins for humans: - Axin-1 - Rhodopsin
Construct a dictionary for these proteins and the number amino acids for each of them. The keys should represent the name of the protein. Display the result.
Now that we have created a dictionary; we can test whether or not a specific key exists our dictionary:
OUTPUT
TrueOUTPUT
FalseUsing in
Using the proteins dictionary you created in the above
challenge, test to see whether or not a protein called
ERK exists as a key in your dictionary?
Display the result as a Boolean value.
Interacting with a dictionary
We have already learnt that in programming, the more explicit our code, the better it is. Interacting with dictionaries in Python is very easy, coherent, and explicit. This makes them a powerful tool that we can exploit for different purposes.
In lists and tuples, we use indexing and
slicing to retrieve values. In dictionaries, however, we use
keys to do that. Because we can define the keys of a
dictionary ourselves, we no longer have to rely exclusively on numeric
indices.
As a result, we can retrieve the values of a dictionary using their respective keys as follows:
OUTPUT
Controls the function of other proteins through phosphorylation.However, if we attempt to retrieve the value for a
key that does not exist in our dict, a
KeyError will be raised:
ERROR
Error in py_call_impl(callable, dots$args, dots$keywords): KeyError: 'GSK3'
Detailed traceback:
File "<string>", line 1, in <module>Dictionary lookup
Implement a dict to represent the following set of
information:
Cystic Fibrosis:
| Full Name | Gene | Type |
|---|---|---|
| Cystic fibrosis transmembrane conductance regulator | CFTR | Membrane Protein |
Using the dictionary you implemented, retrieve and display the gene associated with cystic fibrosis.
Remember
Whilst the values in a dict can be of virtually
any type supported in Python, the keys may only be defined
using immutable types such as string, int, or
tuple. Additionally, the keys in a dictionary must
be unique.
If we attempt to construct a dict using a mutable value
as key, a TypeError will be raised.
For instance, list is a mutable type and therefore
cannot be used as a key:
ERROR
Error in py_call_impl(callable, dots$args, dots$keywords): TypeError: unhashable type: 'list'
Detailed traceback:
File "<string>", line 1, in <module>But we can use any immutable type as a key:
OUTPUT
{'ab': 'some value'}OUTPUT
{('a', 'b'): 'some value'}If we define a key more than once, the Python interpreter
constructs the entry in dict using the last instance.
In the following example, we repeat the key ‘pathway’ twice; and as expected, the interpreter only uses the last instance, which in this case represents the value ‘Canonical’:
PYTHON
signal = {
'name': 'Wnt',
'pathway': 'Non-Canonical', # first instance
'pathway': 'Canonical' # second instance
}
print(signal) {'name': 'Wnt', 'pathway': 'Canonical'}Dictionaries are mutable
Dictionaries are mutable. This means that we can alter their contents. We can make any alterations to a dictionary as long as we use immutable values for the keys.
Suppose we have a dictionary stored in a variable called
protein, holding some information about a specific
protein:
PYTHON
protein = {
'full name': 'Cystic fibrosis transmembrane conductance regulator',
'alias': 'CFTR',
'gene': 'CFTR',
'type': 'Membrane Protein',
'common mutations': ['Delta-F508', 'G542X', 'G551D', 'N1303K']
}We can add new items to our dictionary or alter the existing ones:
OUTPUT
{'full name': 'Cystic fibrosis transmembrane conductance regulator', 'alias': 'CFTR', 'gene': 'CFTR', 'type': 'Membrane Protein', 'common mutations': ['Delta-F508', 'G542X', 'G551D', 'N1303K'], 'chromosome': 7}
7We can also alter an existing value in a dictionary using
its key. To do so, we simply access the value using
its key, and treat it as a normal variable; i.e. the same way
we do with members of a list:
OUTPUT
['Delta-F508', 'G542X', 'G551D', 'N1303K']OUTPUT
{'full name': 'Cystic fibrosis transmembrane conductance regulator', 'alias': 'CFTR', 'gene': 'CFTR', 'type': 'Membrane Protein', 'common mutations': ['Delta-F508', 'G542X', 'G551D', 'N1303K', 'W1282X'], 'chromosome': 7}Altering values
Implement the following dictionary:
signal = {'name': 'Wnt', 'pathway': 'Non-Canonical'}}
with respect to signal:
- Correct the value of pathway to “Canonical”;
- Add a new item to the dictionary to represent the receptors for the canonical pathway as “Frizzled” and “LRP”.
Display the altered dictionary as the final result.
Advanced Topic
Displaying an entire dictionary using the print() function
can look a little messy because it is not properly structured. There is,
however, an external library called pprint (Pretty-Print)
that behaves in very similar way to the default print()
function, but structures dictionaries and other arrays in a more
presentable way before displaying them. We do not discuss
``Pretty-Print’’ in this course, but it is a part of Python’s default
library and is therefore installed with Python automatically. To learn
more it, have a read through the official
documentations for the library and review the examples.
Because the keys are immutable, they cannot be altered. However, we can get around this limitation by introducing a new key and assigning the values of the old key to the new one. Once we do that, we can go ahead and remove the old item. The easiest way to remove an item from a dictionary is to use the syntax del:
PYTHON
# Creating a new key and assigning to it the
# values of the old key:
protein['human chromosome'] = protein['chromosome']
print(protein)OUTPUT
{'full name': 'Cystic fibrosis transmembrane conductance regulator', 'alias': 'CFTR', 'gene': 'CFTR', 'type': 'Membrane Protein', 'common mutations': ['Delta-F508', 'G542X', 'G551D', 'N1303K', 'W1282X'], 'chromosome': 7, 'human chromosome': 7}OUTPUT
{'full name': 'Cystic fibrosis transmembrane conductance regulator', 'alias': 'CFTR', 'gene': 'CFTR', 'type': 'Membrane Protein', 'common mutations': ['Delta-F508', 'G542X', 'G551D', 'N1303K', 'W1282X'], 'human chromosome': 7}We can simplify the above operation using the .pop() method, which removes the specified key from a dictionary and returns any values associated with it:
OUTPUT
{'full name': 'Cystic fibrosis transmembrane conductance regulator', 'alias': 'CFTR', 'gene': 'CFTR', 'type': 'Membrane Protein', 'human chromosome': 7, 'common mutations in caucasians': ['Delta-F508', 'G542X', 'G551D', 'N1303K', 'W1282X']}Reassigning values
Implement a dictionary as:
with respect to signal:
Change the key name from ‘pdb’ to ‘pdb id’ using the .pop() method.
-
Write a code to find out whether the dictionary:
- contains the new key (i.e. ‘pdb id’).
- confirm that it no longer contains the old key (i.e. ‘pdb’)
If both conditions are met, display:
Contains the new key, but not the old one.Otherwise:
Failed to alter the dictionary.Useful methods for dictionary
Now we use some snippets to demonstrate some of the useful
methods associated with dict in Python.
Given a dictionary as:
PYTHON
lac_repressor = {
'pdb id': '1LBI',
'deposit data': '1996-02-17',
'organism': 'Escherichia coli',
'method': 'x-ray',
'resolution': 2.7,
}We can create an array of all items in the dictionary using the .items() method:
OUTPUT
dict_items([('pdb id', '1LBI'), ('deposit data', '1996-02-17'), ('organism', 'Escherichia coli'), ('method', 'x-ray'), ('resolution', 2.7)])The .items() method also returns an array of
tuple members. Each tuple itself consists of 2
members, and is structured as (‘key’: ‘value’). On that account, we
can use its output in the context of a for–loop as
follows:
OUTPUT
pdb id: 1LBI
deposit data: 1996-02-17
organism: Escherichia coli
method: x-ray
resolution: 2.7We learned earlier that if we ask for a key that is not in
the dict, a KeyError will be raised. If we
anticipate this, we can handle it using the .get() method.
The method takes in the key and searches the dictionary to find
it. If found, the associating value is returned. Otherwise, the
method returns None by default. We can also pass a second
value to .get() to replace None in cases that
the requested key does not exist:
OUTPUT
Error in py_call_impl(callable, dots$args, dots$keywords): KeyError: 'gene'
Detailed traceback:
File "<string>", line 1, in <module>OUTPUT
NoneOUTPUT
Not found...Getting multiple values
Implement the lac_repressor dictionary and try to extract the values associated with the following keys:
- organism
- authors
- subunits
- method
If a key does not exist in the dictionary, display No entry instead.
Display the results in the following format:
organism: XXX
authors: XXX PYTHON
lac_repressor = {
'pdb id': '1LBI',
'deposit data': '1996-02-17',
'organism': 'Escherichia coli',
'method': 'x-ray',
'resolution': 2.7,
}
requested_keys = ['organism', 'authors', 'subunits', 'method']
for key in requested_keys:
lac_repressor.get(key, 'No entry')OUTPUT
'Escherichia coli'
'No entry'
'No entry'
'x-ray'
for-loops and dictionaries
Dictionaries and for-loops create a powerful
combination. We can leverage the accessibility of dictionary
values through specific keys that we define ourselves
in a loop to extract data iteratively and repeatedly.
One of the most useful tools that we can create using nothing more
than a for-loop and a dictionary, in only a few lines of
code, is a sequence converter.
Here, we are essentially iterating through a sequence of DNA
nucleotides (sequence),
extracting one character per loop cycle from our string (nucleotide). We then use that
character as a key to retrieve its corresponding value
from our a dictionary (dna2rna). Once we get the
value, we add it to the variable that we initialised using an
empty string outside the scope of our for-loop (rna_sequence). At the end of the
process, the variable rna_sequence will contain a
converted version of our sequence.
PYTHON
sequence = 'CCCATCTTAAGACTTCACAAGACTTGTGAAATCAGACCACTGCTCAATGCGGAACGCCCG'
dna2rna = {"A": "U", "T": "A", "C": "G", "G": "C"}
rna_sequence = str() # Creating an empty string.
for nucleotide in sequence:
rna_sequence += dna2rna[nucleotide]
print('DNA:', sequence)
print('RNA:', rna_sequence)OUTPUT
DNA: CCCATCTTAAGACTTCACAAGACTTGTGAAATCAGACCACTGCTCAATGCGGAACGCCCG
RNA: GGGUAGAAUUCUGAAGUGUUCUGAACACUUUAGUCUGGUGACGAGUUACGCCUUGCGGGCUsing dictionaries as maps
We know that in reverse transcription, RNA nucleotides are converted to their complementary DNA as shown:
| Type | Direction | Nucleotides |
|---|---|---|
| RNA | 5’…’ | U A G C |
| cDNA | 5’…’ | A T C G |
with that in mind:
Use the table to construct a dictionary for reverse transcription, and another dictionary for the conversion of cDNA to DNA.
Using the appropriate dictionary, convert the following mRNA (exon) sequence for human G protein-coupled receptor to its cDNA.
PYTHON
human_gpcr = (
'AUGGAUGUGACUUCCCAAGCCCGGGGCGUGGGCCUGGAGAUGUACCCAGGCACCGCGCAGCCUGCGGCCCCCAACACCACCUC'
'CCCCGAGCUCAACCUGUCCCACCCGCUCCUGGGCACCGCCCUGGCCAAUGGGACAGGUGAGCUCUCGGAGCACCAGCAGUACG'
'UGAUCGGCCUGUUCCUCUCGUGCCUCUACACCAUCUUCCUCUUCCCCAUCGGCUUUGUGGGCAACAUCCUGAUCCUGGUGGUG'
'AACAUCAGCUUCCGCGAGAAGAUGACCAUCCCCGACCUGUACUUCAUCAACCUGGCGGUGGCGGACCUCAUCCUGGUGGCCGA'
'CUCCCUCAUUGAGGUGUUCAACCUGCACGAGCGGUACUACGACAUCGCCGUCCUGUGCACCUUCAUGUCGCUCUUCCUGCAGG'
'UCAACAUGUACAGCAGCGUCUUCUUCCUCACCUGGAUGAGCUUCGACCGCUACAUCGCCCUGGCCAGGGCCAUGCGCUGCAGC'
'CUGUUCCGCACCAAGCACCACGCCCGGCUGAGCUGUGGCCUCAUCUGGAUGGCAUCCGUGUCAGCCACGCUGGUGCCCUUCAC'
'CGCCGUGCACCUGCAGCACACCGACGAGGCCUGCUUCUGUUUCGCGGAUGUCCGGGAGGUGCAGUGGCUCGAGGUCACGCUGG'
'GCUUCAUCGUGCCCUUCGCCAUCAUCGGCCUGUGCUACUCCCUCAUUGUCCGGGUGCUGGUCAGGGCGCACCGGCACCGUGGG'
'CUGCGGCCCCGGCGGCAGAAGGCGCUCCGCAUGAUCCUCGCGGUGGUGCUGGUCUUCUUCGUCUGCUGGCUGCCGGAGAACGU'
'CUUCAUCAGCGUGCACCUCCUGCAGCGGACGCAGCCUGGGGCCGCUCCCUGCAAGCAGUCUUUCCGCCAUGCCCACCCCCUCA'
'CGGGCCACAUUGUCAACCUCACCGCCUUCUCCAACAGCUGCCUAAACCCCCUCAUCUACAGCUUUCUCGGGGAGACCUUCAGG'
'GACAAGCUGAGGCUGUACAUUGAGCAGAAAACAAAUUUGCCGGCCCUGAACCGCUUCUGUCACGCUGCCCUGAAGGCCGUCAU'
'UCCAGACAGCACCGAGCAGUCGGAUGUGAGGUUCAGCAGUGCCGUG'
)Q1:
PYTHON
mrna2cdna = {
'U': 'A',
'A': 'T',
'G': 'C',
'C': 'G'
}
cdna2dna = {
'A': 'T',
'T': 'A',
'C': 'G',
'G': 'C'
}Q2:
OUTPUT
TACCTACACTGAAGGGTTCGGGCCCCGCACCCGGACCTCTACATGGGTCCGTGGCGCGTCGGACGCCGGGGGTTGTGGTGGAGGGGGCTCGAGTTGGACAGGGTGGGCGAGGACCCGTGGCGGGACCGGTTACCCTGTCCACTCGAGAGCCTCGTGGTCGTCATGCACTAGCCGGACAAGGAGAGCACGGAGATGTGGTAGAAGGAGAAGGGGTAGCCGAAACACCCGTTGTAGGACTAGGACCACCACTTGTAGTCGAAGGCGCTCTTCTACTGGTAGGGGCTGGACATGAAGTAGTTGGACCGCCACCGCCTGGAGTAGGACCACCGGCTGAGGGAGTAACTCCACAAGTTGGACGTGCTCGCCATGATGCTGTAGCGGCAGGACACGTGGAAGTACAGCGAGAAGGACGTCCAGTTGTACATGTCGTCGCAGAAGAAGGAGTGGACCTACTCGAAGCTGGCGATGTAGCGGGACCGGTCCCGGTACGCGACGTCGGACAAGGCGTGGTTCGTGGTGCGGGCCGACTCGACACCGGAGTAGACCTACCGTAGGCACAGTCGGTGCGACCACGGGAAGTGGCGGCACGTGGACGTCGTGTGGCTGCTCCGGACGAAGACAAAGCGCCTACAGGCCCTCCACGTCACCGAGCTCCAGTGCGACCCGAAGTAGCACGGGAAGCGGTAGTAGCCGGACACGATGAGGGAGTAACAGGCCCACGACCAGTCCCGCGTGGCCGTGGCACCCGACGCCGGGGCCGCCGTCTTCCGCGAGGCGTACTAGGAGCGCCACCACGACCAGAAGAAGCAGACGACCGACGGCCTCTTGCAGAAGTAGTCGCACGTGGAGGACGTCGCCTGCGTCGGACCCCGGCGAGGGACGTTCGTCAGAAAGGCGGTACGGGTGGGGGAGTGCCCGGTGTAACAGTTGGAGTGGCGGAAGAGGTTGTCGACGGATTTGGGGGAGTAGATGTCGAAAGAGCCCCTCTGGAAGTCCCTGTTCGACTCCGACATGTAACTCGTCTTTTGTTTAAACGGCCGGGACTTGGCGAAGACAGTGCGACGGGACTTCCGGCAGTAAGGTCTGTCGTGGCTCGTCAGCCTACACTCCAAGTCGTCACGGCAC