# fix incorrect values # old value # new value
linelist <- linelist %>%
mutate(date_onset = recode(date_onset, "2014-14-15" = "2014-04-15"))Recoding Values
You can find the original, extended version of this chapter here.
Here are a few scenarios where you need to re-code (change) values:
- to edit one specific value (e.g. one date with an incorrect year or format)
- to reconcile values not spelled the same
- to create a new column of categorical values
- to create a new column of numeric categories (e.g. age categories)
Manually changing specific values
To change values manually you can use the recode() function within the mutate() function.
Imagine there is a nonsensical date in the data (e.g. “2014-14-15”): you could fix the date manually in the raw source data, or, you could write the change into the cleaning pipeline via mutate() and recode(). The latter is more transparent and reproducible to anyone else seeking to understand or repeat your analysis.
The mutate() line above can be read as: “mutate the column date_onset to equal the column date_onset re-coded so that OLD VALUE is changed to NEW VALUE”. Note that this pattern (OLD = NEW) for recode() is the opposite of most R patterns (new = old). The R development community is working on revising this.
By logic
Below we demonstrate how to re-code values in a column using logic and conditions:
- Using
replace(),ifelse()andif_else()for simple logic - Using
case_when()for more complex logic
Simple logic
replace()
To re-code with simple logical criteria, you can use replace() within mutate(). replace() is a function from base R. Use a logic condition to specify the rows to change . The general syntax is:
mutate(col_to_change = replace(col_to_change, criteria for rows, new value)).
One common situation to use replace() is changing just one value in one row, using an unique row identifier. Below, the gender is changed to “Female” in the row where the column case_id is “2195”.
# Example: change gender of one specific observation to "Female"
linelist <- linelist %>%
mutate(gender = replace(gender, case_id == "2195", "Female"))The equivalent command using base R syntax and indexing brackets [ ] is below. It reads as “Change the value of the dataframe linelist‘s column gender (for the rows where linelist’s column case_id has the value ’2195’) to ‘Female’”.
linelist$gender[linelist$case_id == "2195"] <- "Female"ifelse() and if_else()
Another tool for simple logic is ifelse() and its partner if_else(). However, in most cases for re-coding it is more clear to use case_when() (detailed below). These “if else” commands are simplified versions of an if and else programming statement. The general syntax is:
ifelse(condition, value to return if condition evaluates to TRUE, value to return if condition evaluates to FALSE)
Below, the column source_known is defined. Its value in a given row is set to “known” if the row’s value in column source is not missing. If the value in source is missing, then the value in source_known is set to “unknown”.
linelist <- linelist %>%
mutate(source_known = ifelse(!is.na(source), "known", "unknown"))if_else() is a special version from dplyr that handles dates. Note that if the ‘true’ value is a date, the ‘false’ value must also qualify a date, hence using the special value NA_real_ instead of just NA.
# Create a date of death column, which is NA if patient has not died.
linelist <- linelist %>%
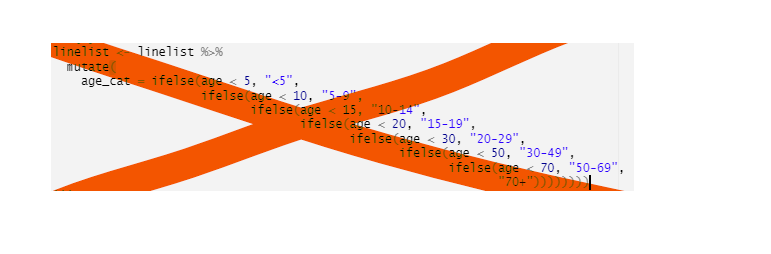
mutate(date_death = if_else(outcome == "Death", date_outcome, NA_real_))Avoid stringing together many ifelse commands… use case_when() instead! case_when() is much easier to read and you’ll make fewer errors.
Outside of the context of a data frame, if you want to have an object used in your code switch its value, consider using switch() from base R.
Complex logic
Use dplyr’s case_when() if you are re-coding into many new groups, or if you need to use complex logic statements to re-code values. This function evaluates every row in the data frame, assess whether the rows meets specified criteria, and assigns the correct new value.
case_when() commands consist of statements that have a Right-Hand Side (RHS) and a Left-Hand Side (LHS) separated by a “tilde” ~. The logic criteria are in the left side and the pursuant values are in the right side of each statement. Statements are separated by commas.
For example, here we utilize the columns age and age_unit to create a column age_years:
linelist <- linelist %>%
mutate(age_years = case_when(
age_unit == "years" ~ age, # if age unit is years
age_unit == "months" ~ age/12, # if age unit is months, divide age by 12
is.na(age_unit) ~ age)) # if age unit is missing, assume years
# any other circumstance, assign NA (missing)As each row in the data is evaluated, the criteria are applied/evaluated in the order the case_when() statements are written - from top-to-bottom. If the top criteria evaluates to TRUE for a given row, the RHS value is assigned, and the remaining criteria are not even tested for that row in the data. Thus, it is best to write the most specific criteria first, and the most general last. A data row that does not meet any of the RHS criteria will be assigned NA.
Sometimes, you may with to write a final statement that assigns a value for all other scenarios not described by one of the previous lines. To do this, place TRUE on the left-side, which will capture any row that did not meet any of the previous criteria. The right-side of this statement could be assigned a value like “check me!” or missing.
Below is another example of case_when() used to create a new column with the patient classification, according to a case definition for confirmed and suspect cases:
linelist <- linelist %>%
mutate(case_status = case_when(
# if patient had lab test and it is positive,
# then they are marked as a confirmed case
ct_blood < 20 ~ "Confirmed",
# given that a patient does not have a positive lab result,
# if patient has a "source" (epidemiological link) AND has fever,
# then they are marked as a suspect case
!is.na(source) & fever == "yes" ~ "Suspect",
# any other patient not addressed above
# is marked for follow up
TRUE ~ "To investigate"))Numeric categories
Here we describe some special approaches for creating categories from numerical columns. Common examples include age categories, groups of lab values, etc. Here we will discuss:
age_categories(), from the epikit package
cut(), from base R
case_when()
- quantile breaks with
quantile()andntile()
Review distribution
For this example we will create an age_cat column using the age_years column.
#check the class of the linelist variable age
class(linelist$age_years)[1] "numeric"cut()
The basic syntax within cut() is to first provide the numeric column to be cut (age_years), and then the breaks argument, which is a numeric vector c() of break points. Using cut(), the resulting column is an ordered factor.
By default, the categorization occurs so that the right/upper side is “open” and inclusive (and the left/lower side is “closed” or exclusive). This is the opposite behavior from the age_categories() function. The default labels use the notation “(A, B]”, which means A is not included but B is. Reverse this behavior by providing the right = TRUE argument.
Thus, by default, “0” values are excluded from the lowest group, and categorized as NA! “0” values could be infants coded as age 0 so be careful! To change this, add the argument include.lowest = TRUE so that any “0” values will be included in the lowest group. The automatically-generated label for the lowest category will then be “[A],B]”. Note that if you include the include.lowest = TRUE argument and right = TRUE, the extreme inclusion will now apply to the highest break point value and category, not the lowest.
You can provide a vector of customized labels using the labels = argument. As these are manually written, be very careful to ensure they are accurate! Check your work using cross-tabulation, as described below.
An example of cut() applied to age_years to make the new variable age_cat is below:
# Create new variable, by cutting the numeric age variable
# lower break is excluded but upper break is included in each category
linelist <- linelist %>%
mutate(
age_cat = cut(
age_years,
breaks = c(0, 5, 10, 15, 20,
30, 50, 70, 100),
include.lowest = TRUE # include 0 in lowest group
))
# tabulate the number of observations per group
table(linelist$age_cat, useNA = "always")
[0,5] (5,10] (10,15] (15,20] (20,30] (30,50] (50,70] (70,100]
1315 1065 930 696 1013 694 84 5
<NA>
86 Check your work!!! Verify that each age value was assigned to the correct category by cross-tabulating the numeric and category columns. Examine assignment of boundary values (e.g. 15, if neighboring categories are 10-15 and 16-20).
Quantile breaks
In common understanding, “quantiles” or “percentiles” typically refer to a value below which a proportion of values fall. For example, the 95th percentile of ages in linelist would be the age below which 95% of the age fall.
However in common speech, “quartiles” and “deciles” can also refer to the groups of data as equally divided into 4, or 10 groups (note there will be one more break point than group).
To get quantile break points, you can use quantile() from the stats package from base R. You provide a numeric vector (e.g. a column in a dataset) and vector of numeric probability values ranging from 0 to 1.0. The break points are returned as a numeric vector. Explore the details of the statistical methodologies by entering ?quantile.
- If your input numeric vector has any missing values it is best to set
na.rm = TRUE
- Set
names = FALSEto get an un-named numeric vector
quantile(linelist$age_years, # specify numeric vector to work on
probs = c(0, .25, .50, .75, .90, .95), # specify the percentiles you want
na.rm = TRUE) # ignore missing values 0% 25% 50% 75% 90% 95%
0.0 6.0 13.0 23.0 33.9 41.0 You can use the results of quantile() as break points in age_categories() or cut(). Below we create a new column deciles using cut() where the breaks are defined using quantiles() on age_years. Below, we display the results using tabyl() from janitor so you can see the percentages. Note how they are not exactly 10% in each group.
linelist %>% # begin with linelist
mutate(deciles = cut(age_years, # create new column decile as cut() on column age_years
breaks = quantile( # define cut breaks using quantile()
age_years, # operate on age_years
probs = seq(0, 1, by = 0.1), # 0.0 to 1.0 by 0.1
na.rm = TRUE), # ignore missing values
include.lowest = TRUE)) %>% # for cut() include age 0
janitor::tabyl(deciles) # pipe to table to display deciles n percent valid_percent
[0,2] 658 0.11175272 0.11340917
(2,5] 657 0.11158288 0.11323681
(5,7] 447 0.07591712 0.07704240
(7,10] 618 0.10495924 0.10651499
(10,13] 572 0.09714674 0.09858669
(13,17] 674 0.11447011 0.11616684
(17,21] 520 0.08831522 0.08962427
(21,26] 547 0.09290082 0.09427784
(26,33.9] 528 0.08967391 0.09100310
(33.9,84] 581 0.09867527 0.10013788
<NA> 86 0.01460598 NA